This is an old revision of the document!
Table of Contents
Back to Manny's Bioinformatics Workshop Home
What is Linux ?
It is a free and open source operating system released in 1991 under the GNU GPL license.GPL allows anyone to use, modify and redistribute with the requirement that they pass it on with the same license.
It is the leading operating system of choice for servers such as supercomputers. More than 90% of the top 500 fastest computers are based on Linux.
MAC computers are related to Linux because they are also based on UNIX
Depending on the purpose of the Linux machine, it may or may not have a Desktop environment that we are familiar with on our personal computers. Linux uses X Window System to provide the Desktop environment.
A popular distribution of Linux operating system is called Ubuntu.
Why do bioinformaticians use Linux?
- Many bioinformatics core tools are written in Linux.
- BLAST, CLUSTALW, PHRAP, etc
- Many web applications are also supported on web servers hosted on linux machines
- Linux supports development of software for many different programming languages.
- Developers are lazy so creating a software that does not require a window is much faster and easier
- Multiple users can log in at the same time.
- A user logging in over the network can do just about anything a user sitting in front of the computer can do. Which also means linux handles multitasking very well.
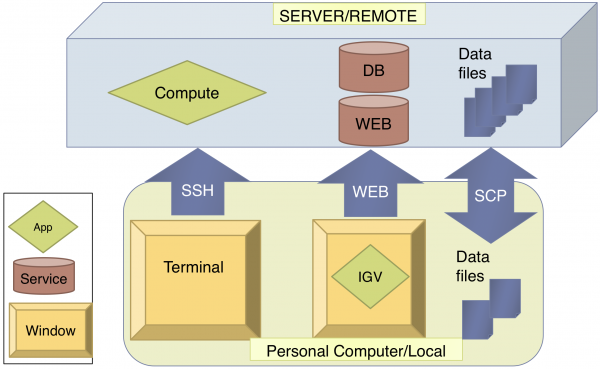
Remote vs. Local
The Linux Shell
The standard user interface for personal computers is a GUI (Graphical User Interface). However for linux it is a command-line interpreter called shell. It is simply a prompt the awaits your command. There are several different shells, but the one used often is called “bash”, which is a mixture of a bunch of other shells.
Command Line Editing
The command is only executed once you press enter. Till then you can edit the line by using the following key strokes:
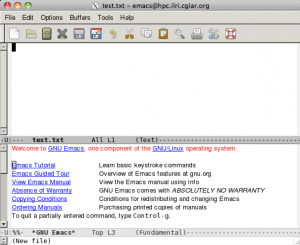
Logging in with X Windows
In cases where a program requires a GUI, you should log in using the –X option. This opens a tunnel to your computer allowing all windows to open in your computer. For this to work you need X11 installed on your computer (MobaXterm already has one) MAC – Xquartz (http://xquartz.macosforge.org/landing/) Windows – Xming (http://sourceforge.net/projects/xming/)
Last login: Wed Jun 3 15:49:01 on ttys000 Manpreets-MacBook-Pro:~ manpreetkatari$ ssh mkatari@hpc.ilri.cgiar.org Unauthorized access is prohibited. mkatari@hpc.ilri.cgiar.org's password: Last login: Wed Jun 3 16:33:26 2015 from 197.136.62.11 [mkatari@hpc ~]$
You should have a window popup on your computer that looks something like this.
Some useful information about linux
Environment variables and PATH
All variables that are set in your environment can be found by using
env
The variable that is most important to us is PATH. The PATH is where the computer is looking for the commands. To see the contents of the variable type:
echo $PATH
In the sbatch files we have been adding the full path to the commands. Another option is to add the full path of the command you want to use to the variable PATH. This is what module load essentially does.
For example the following two commands are equivalent.
export PATH=/export/apps/samtools/0.1.19/bin:$PATH module load samtools